LASERGENE 17.2 RELEASE NOTES
Lasergene 17.2 is now available to download. This release incorporates your feedback to improve the workflows for Sanger sequence assemblies, NGS assemblies and variant analysis, and multiple sequence alignments.
Updates and Improvements for Sanger Sequence Assemblies (SeqMan Ultra and SeqMan NGen)
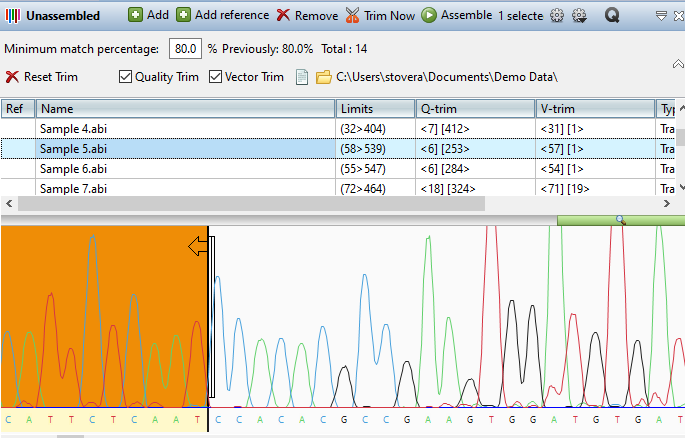
- Based on your feedback, we’ve significantly improved the workflow experience for Sanger sequence assemblies by fully integrating SeqMan NGen into SeqMan Ultra. This includes adding a new Unassembled Sequences window to SeqMan Ultra, which allows you to preview and trim trace data prior to assembly.
- We’ve also made vector trimming easier by using our extensive vector database by default. As always, you can also add your own vectors.
- Our new search toolbar makes it simple to search individual sequences or all sequences in your assembly by coverage, name, or specific bases.
- Easily export an image of your assembly for collaboration or publication in PDF, Bitmap, or PPT format.
- We’ve added a new Majority Consensus Track to make it easier to compare the Majority method to our proprietary and more accurate Trace Consensus caller.
- We’ve also added a new Reference Track to avoid confusion that sometimes happens with differing coordinates between the reference and consensus sequence.
- Conveniently manage and access all of your recent assemblies from the new Jobs panel.
Updates and Improvements for NGS Assembly and Analysis (Lasergene Genomics)
- Based on your feedback, we’ve fully integrated SeqMan NGen into SeqMan Ultra for NGS assemblies so that you can go from our streamlined project setup to finished assembly all in one application. You can still set up your project directly in SeqMan NGen, if you prefer.
- Our Variant Annotation Database, included in Lasergene Genomics, is now integrated with Mastermind, the comprehensive database of genomic literature from Genomenon. With this integration, Lasergene users can now quickly and easily search and cross-reference NGS variant data from millions of PubMed publications.
- We’ve also updated our comprehensive Variant Annotation Database to include dbNSFP version 4.1, as well as allele and gene frequencies.
- A new unzipping tool has been incorporated into SeqMan NGen so that your zipped .fastq input files are conveniently converted automatically.
- Conveniently manage and access all of your recent assemblies from the new Jobs panel in SeqMan Ultra.
New Features and Analysis Options for Multiple Sequence Alignments and Phylogenetic Trees (MegAlign Pro)
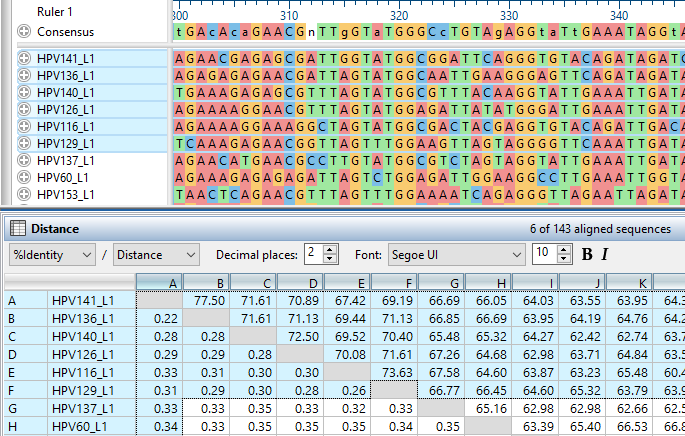
- The Distance Table for multiple sequence alignments is now more robust and flexible. Now both % Identity and Distance are displayed by default, and we’ve integrated several new statistical options to evaluate for your data. We’ve also added more convenient access to adjust parameters, and a new quick style bar to easily apply statistics and formatting.
- The new Styles feature in this release allows you to save default styles, such as residue color. We’ve made it easy to save multiple styles, so you can have different default settings for nucleotide and amino acid sequences, for example. This complements the Layouts feature, which allows you to save how document views are arranged by default.
- We’ve added a new sorting feature, giving you the ability to organize sequences on the phylogenetic tree by depth or distance, or to sort sequences in the alignment to reflect the order of sequences in the tree.
- Our new easy access search feature makes it simple to search and navigate to specific sequences by sequence name.



Leave a Reply
Your email is safe with us.